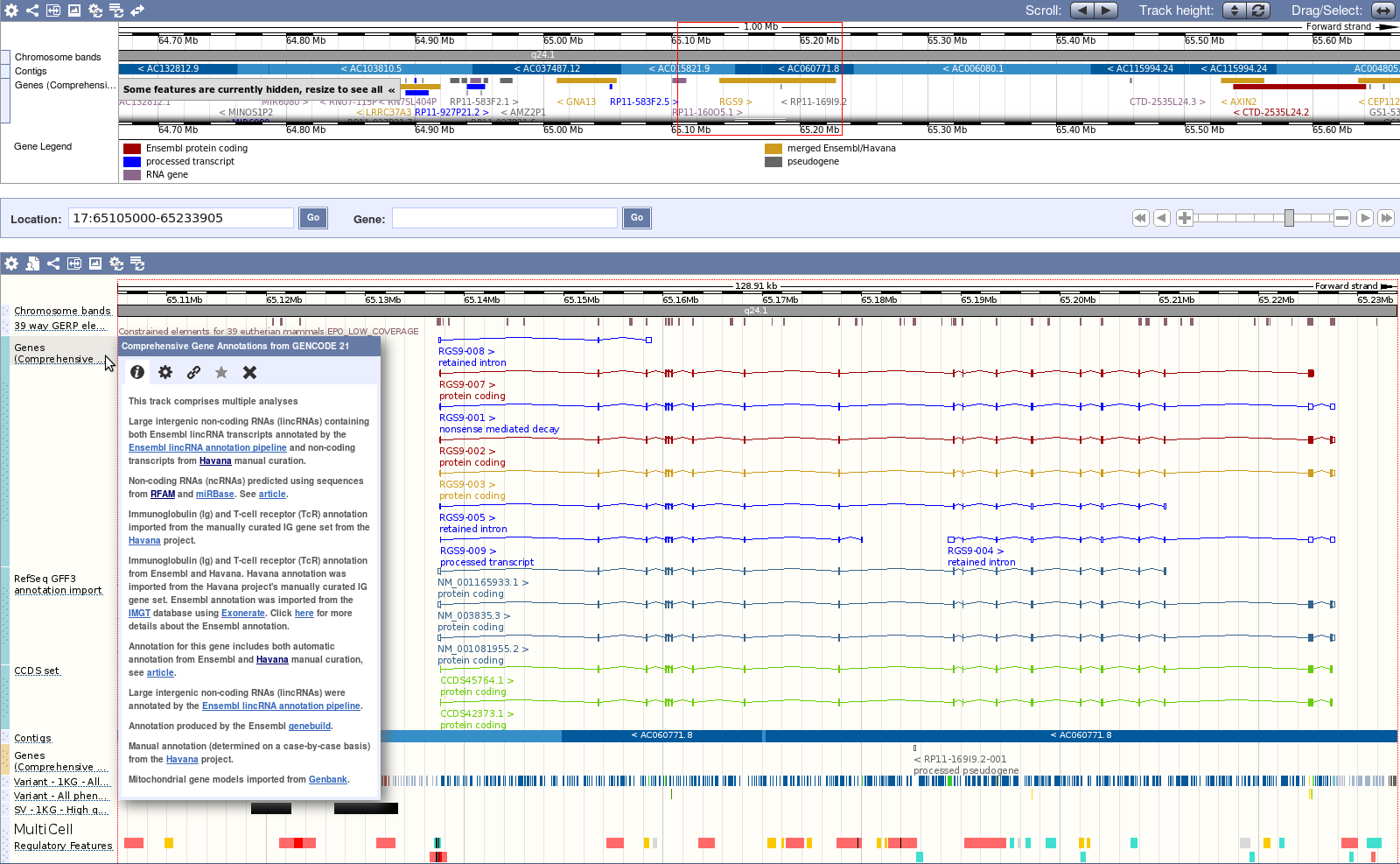
How to access the data
All the GENCODE project data is open access and can be accessed by any of the following methods. If you need help, please contact us.
- The GENCODE release files can be found in this website or directly in our human and mouse FTP sites.
-
The current version of GENCODE is the default geneset in the Ensembl Genome Browser for human and mouse.
The Ensembl data can also be accessed using the options listed below:
-
The UCSC Genome Browser hosts selected GENCODE releases. GENCODE track data can also be downloaded from the Table Browser.
The links below can be used to add GENCODE-related track hubs in the Ensembl or UCSC Genome Browsers:
- GENCODE annotation updates: manual annotation updated since the last release.
- GENCODE annotation releases: human releases since v7 and all mouse releases.
- RACE-454seq on GENCODE lncRNAs: extension of human lncRNA transcripts by RACE coupled with long read high-throughput sequencing.
- GENCODE CLS CaptureSeq: Capture Long-Seq (CLS) on human and mouse lincRNAs.
- PhyloCSF: protein-coding potential based on comparative genomics.
- We have created a custom long non-coding RNA expression array design with probes targeting the Gencode v15 human lncRNA annotation.